v2306: New and improved parallel operation
An extension to the parallel profiling function object, parProfiling, limits profiling to a linear solver. The function object is specified in the controlDict using, e.g.
functions
{
// Run parProfiling
profiling
{
libs (utilityFunctionObjects);
#includeEtc "caseDicts/profiling/parallel.cfg"
detail 2;
}
}
It takes the form of a new parProfiling wrapper solver in the fvSolution file:
solvers
{
p
{
solver parProfiling;
baseSolver PCG;
preconditioner DIC;
tolerance 1e-06;
relTol 0.05;
}
}
The example above enables the parProfiling function object only for the PCG linear solver. This extension is particularly useful when looking at the effect of the coarsest-level linear solver inside the GAMG solver.
A typical output:
parProfiling:
reduce : avg = 72.7133s
min = 2.37s (processor 0)
max = 88.29s (processor 4)
all-all : avg = 14.9633s
min = 11.04s (processor 5)
max = 17.18s (processor 4)
A combination of both is very useful to investigate the effect of different linear solvers, e.g. with the cavity lid-driven tutorial on 20 processors:
parProfiling output for PCG:
reduce : ..
..
counts 20(14875 14875 14875 14875 14875 14875 14875 14875 14875 14875 14875 14875 14875 14875 14875 14875 14875 14875 14875 14875)
parProfiling output for FPCG:
reduce : ..
..
counts 20(11914 11914 11914 11914 11914 11914 11914 11914 11914 11914 11914 11914 11914 11914 11914 11914 11914 11914 11914 11914)
Source code
Tutorial
Merge request
The multiLevel decomposition method can be used to preferentially limit the amount of inter-node cuts (uses slow communication) compared to the intra-node (uses faster communication). This functionality now exists natively in the scotch (not ptscotch) decomposition method by specifying a set of weights:
method scotch;
numberOfSubdomains 2048;
coeffs
{
// Divide into 64 nodes, each of 32 cores
domains (64 32);
// Inside a nodes the communication weight is 1% of that inbetween nodes
domainWeights (1 0.01);
}
Alternatively, the first-level decomposition can be omitted by assuming weights are 1 for that level:
method scotch;
numberOfSubdomains 2048;
coeffs
{
// Divide into 2048/32=64 nodes
domains (32);
// Inside a node the communication weight is 1% of that inbetween nodes
domainWeights (0.01);
}
The image below shows the result of decomposing a simple 20x20 mesh into 3 sections of each 32:
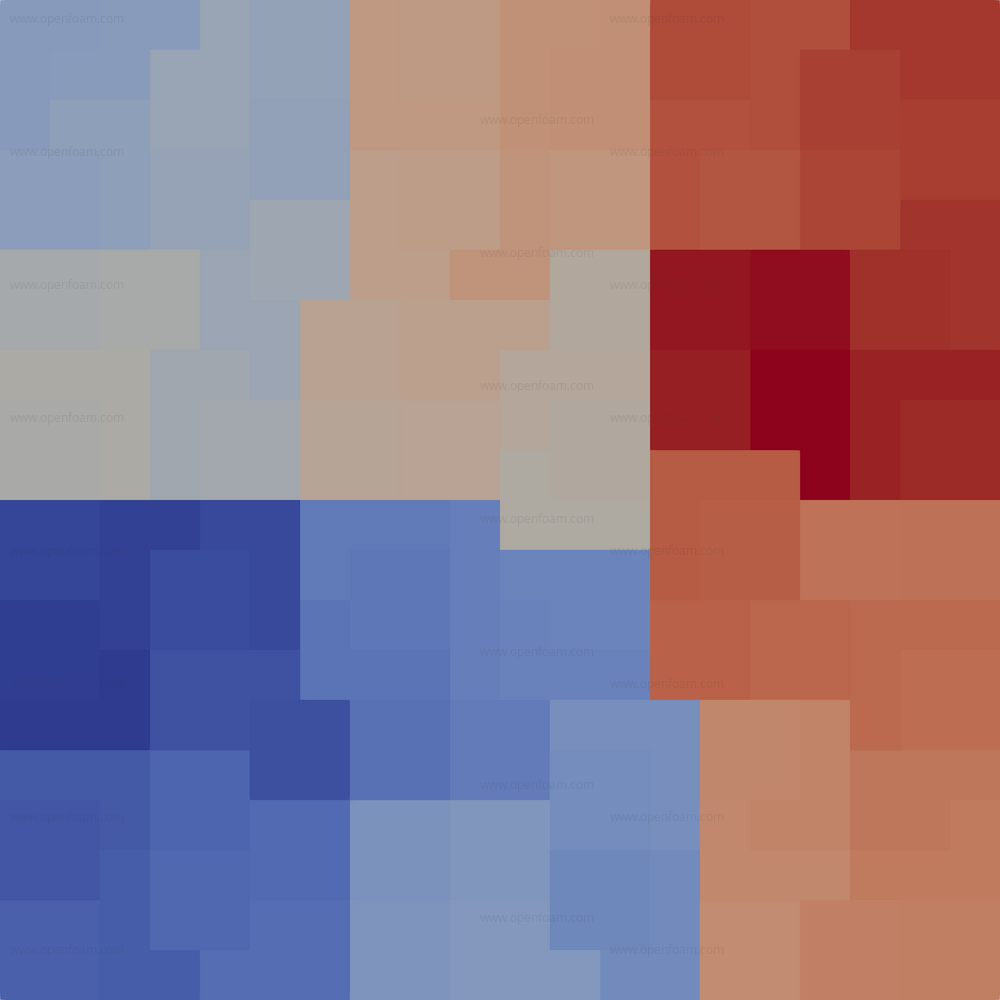
Compared to the multiLevel method it
- is much faster - multiLevel subsets each first level decomposition to decompose the second level. For higher number of cells this becomes a bottleneck.
- avoids creating excessive numbers of processor patches as occasionally happens with the multiLevel method.
Unfortunately the functionality is not implemented in the external ptscotch library; once available it is trivial to add it to the ptscotch decomposition method.
Source code
Merge request
When running in parallel the linear solver waits for all communication to finish before the boundary conditions ('interfaces') are updated using the received data. This can cause a bottleneck when one of the communications is slower than the others, e.g. maybe a slower connection or more faces in the interface.
To avoid this, the nPollProcInterfaces optimisation switch was introduced to enable polling of the interfaces and perform the interface update as they become 'ready'. This switch is either a fixed number of iterations (followed by a blocking wait) or '-1' for polling only (new functionality):
In the etc/controlDict or the local system/controlDict set the nPollProcInterfaces to -1:
OptimisationSwitches
{
// Fixed or infinite (-1) number of polling iterations
nPollProcInterfaces -1;
}
As a general rule:
- Use when there is enough local work, i.e. adding to the solution for processor interfaces, that it makes sense to overlap with communication. Note that this is not the case for the coarsest level in GAMG - there are too few cells.
- Use when there is a massive difference between minimum and maximum processor interface sizes, i.e. numbers of faces. This might be the case for, e.g. geometric decomposition methods, but not generally for topological decomposition methods, e.g. scotch.
A big disadvantage is that the order of additions to the solution is now non-deterministic and this will affect the truncation error, e.g. running the pitzDaily incompressible tutorial with a (bad) solver combination:
p
{
solver PCG;
preconditioner diagonal;
tolerance 1e-06;
relTol 0.1;
}
produces a noticeable solution difference after 100 iterations:
- no polling:
diagonalPCG: Solving for p, Initial residual = 0.012452, Final residual = 0.0012432, No Iterations 162
- polling:
diagonalPCG: Solving for p, Initial residual = 0.012544, Final residual = 0.0012536, No Iterations 301
When running GAMG on many cores the coarsest level solver, e.g. PCG, might become the scaling bottleneck. One way to get around this is to agglomerate coarse level matrices onto less or even one processor. This collects processors' matrices to create a single, larger matrix with all the inter-processor boundaries replaced with internal faces. This avoids communication but adds additional computational cost on the processor that solves this agglomerated matrix. If, e.g. the original agglomeration was down to 1 cell on 10000 processors, the agglomerated matrix will have at least 10000 cells---solving this might now become the bottleneck.
In this development the masterCoarsest processor agglomeration can re-start the local agglomeration to take those 10000 cells back to less. The new behaviour is triggered by a new parameter nCellsInMasterLevel in the fvSolution, e.g.
p
{
solver GAMG;
..
smoother GaussSeidel;
//- Local agglomeration parameters
nCellsInCoarsestLevel 1;
//- Processor-agglomeration parameters
processorAgglomerator masterCoarsest;
//- Optionally agglomerate after processor-agglomeration
nCellsInMasterLevel 1;
}
Results
As a simple testcase the pitzDaily tutorial was modified to run in parallel on 15 processors (using scotch as a decomposition method). With masterCoarsest but no restarting of agglomeration the first iteration will show
DICPCG: Solving for coarsestLevelCorr, Initial residual = 1, Final residual = 0.0788583, No Iterations 6
DICPCG: Solving for coarsestLevelCorr, Initial residual = 1, Final residual = 0.0150588, No Iterations 6
..
DICPCG: Solving for coarsestLevelCorr, Initial residual = 1, Final residual = 0.0105486, No Iterations 6
GAMG: Solving for p, Initial residual = 1, Final residual = 0.0759038, No Iterations 24
With nCellsInMasterLevel 1:
DICPCG: Solving for coarsestLevelCorr, Initial residual = 1, Final residual = 0, No Iterations 1
DICPCG: Solving for coarsestLevelCorr, Initial residual = 1, Final residual = 1.11661e-16, No Iterations 1
..
DICPCG: Solving for coarsestLevelCorr, Initial residual = 1, Final residual = 0, No Iterations 1
GAMG: Solving for p, Initial residual = 1, Final residual = 0.0799836, No Iterations 24
Source code
Merge request
Findings from recent exaFOAM activities have led to many low-level improvements to the OpenFOAM Pstream library (MPI interface) as well as a number of algorithmic and code changes to reduce communication bottlenecks across the code base. The effect of most of these changes will, however, only be evident on larger scale simulations. Since some of the algorithmic changes are considered as *preview* changes, they are not enabled by default but via configuration switches.
For code developers, the handling of MPI requests and communicators has been extended to allow more flexibility when writing your own parallel algorithms. Handling of size exchanges using a non-blocking consensus exchange (NBX) is provided as a drop-in replacement for an all-to-all exchange.

