OpenFOAM® v1606+: New Parallel Functionality
Performance improvements
Parallel scalability tests indicate that OPENFOAM® generally performs well on larger numbers of processors, except for some bottlenecks that are particularly visible in certain solvers.
Following a review by the Japanese Research Organisation for Information Science and Technology (RIST) a collection of enhancements were suggested, based on tests using the pimpleDyMFoam solver. The resulting recommendations lead to a significant performance benefit when employing a large number of processors. The optimisations comprised:
- all-to-all processor size information exchange (performance)
- changing the gather/scatter order (performance)
- on-the-fly calculation of the communication schedule (memory)
All-to-all size information exchange
In some code phases, e.g. mesh geometry calculations, Lagrangian particle exchange,
OPENFOAM® uses a parallel exchange to determine the size
of data to be sent, and uses this to make decisions and pre-size the buffers for the
data to be received. In earlier OPENFOAM® versions, every
processor distributed the size to be sent not only to itself, but also to all other
processors, i.e. all processors would collect a table of size 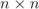 where
where  is the
number of processors and for larger numbers of processors this incurs a large
communications overhead.
is the
number of processors and for larger numbers of processors this incurs a large
communications overhead.
In this version we only obtain the sizes for the current processor. This improves the performance of any variable-sized parallel transfer e.g. geometry initialisation, geometry overlap calculation, Lagrangian particle transfer. This leads to a substantial improvement for moving mesh solvers. For non-Lagrangian, static mesh solvers the effect is a slightly reduced startup time.
Changing the gather/scatter order
Parallel gather/scatter routines are used to synchronise data across processors. Those used most often e.g. in the linear solvers, are already heavily optimised but all others are coded as a gather followed by a scatter operation. These routines use ‘blocking’ communication so as not to allocate any buffers (since the amount to send is generally trivial) and a tree-like communication schedule where each processor receives from ‘below’ processors and sends to the ‘above’ processor (and vice-versa for scatter) The tree is asymmetric such that in ‘gather’ the order of receive is such that the ‘below’ processor with the lowest sub-tree depth is received from first, thus minimising the critical path. In this version a similar optimisation has been done for the ‘scatter’/sending algorithm. Unlike the previous optimisation, this affects all solvers.
![[Picture]](/releases/openfoam-v1606/img/parallel-performance-improvements-small.png)
On-the-fly calculation of the communication schedule
This is a memory optimisation. Above communication schedule gets calculated on all processors and is global i.e. the same on all processors. Hence it can consume an extremely large amount of memory on large clusters. This optimisation calculates only that part of the schedule that is needed. On very large clusters (tested using 6144 cores) there can be more than a halving of the maximum memory usage.
The net effect of these optimisations on a medium sized cluster with a fast network, e.g. 128 processors, 6 cores per node, DDR Infiniband network, shows an improvement of approximately 5% when using the rhoPimpleDyMFoam solver,
![[Picture]](/releases/openfoam-v1606/img/parallel-performance-improvements-rhopimpledymfoam-small.png)
and similar values for the rhoPimpleFoam solver. The amount of speedup expected scales roughly with the square of the number of processors, suggesting significant benefits at higher processor counts.
- Acknowledgement
- Thanks to Mr. Y. Inoue (RIST) for the analysis and suggested improvements to OpenFOAM.
Updated parallel utilities
decomposePar, reconstructPar, redistributePar: These utilities have been extended to decompose/reconstruct the refinement data generated by snappyHexMesh and dynamic (un)refinement. The enables e.g. runs with dynamic refinement/unrefinement to be continued with a different number of processors.
The only limitation is that unrefinement (undoing of refinement) can only happen if all the refined cells are on the same processor. A new decomposition constraint, refinementHistory, can be used to enforce this. It keeps all cells originating from a single cell on the same processor and thus constrains the decomposition such that the refinement can be completely undone (at the cost of potentially a small imbalance)

